Conditional average treatment effect estimation
Source:vignettes/ConditionalAverageTreatmentEffect.Rmd
ConditionalAverageTreatmentEffect.Rmd
require(glmnet)
#> Loading required package: glmnet
#> Loading required package: Matrix
#> Loaded glmnet 4.1-10Background: CATE estimation and pretraining
In causal inference, we are often interested in predicting the treatment effect for individual observations; this is called the conditional average treatment effect (CATE). For example, before prescribing a drug to a patient, we want to know whether the drug is likely to work well for that patient - not just whether it works well on average. One tool to model the CATE is the R-learner (Nie and Wager (2021)), which minimizes the R loss:
$$ \hat{L}_n\{\tau(\cdot)\}=\arg \min_\tau \frac{1}{n}\sum\Bigl[ (y_i- m^*(x_i)) - (W_i-e^*(x_i))\tau(x_i) \Bigr]^2. $$ Here, and are the covariates and outcome for observation , is the treatment propensity and the treatment assignment, and is the conditional mean outcome (). Then, is the estimate of the heterogeneous treatment effect function.
This is fitted in stages: first, the R-learner fits and to get and ; then plugs in and to fit . A minor detail is that cross-fitting (or prevalidation) is used in the first stage so that the plugin value for e.g. comes from a model trained without using .
When is a linear function, then the second stage of fitting is straightforward. The values and are known, and we can use linear regression to model as a function of the weighted feature vector . This is what we will do in the following example.
How can pretraining be useful here? Well, we are separately fitting models for (the conditional mean) and (the heterogeneous treatment effect), and these two functions are likely to share support: it is sensible to assume that the features that modulate the mean treatment effect also modulate the heterogeneous treatment effect. We can use pretraining by (1) training a model for and (2) using the support from this model to guide the fitting of . Note that the offset is not used in this case; and are designed to predict different outcomes.
A simulated example
Here is an example. We will simplify the problem by assuming treatment has been randomized – the true for all .
set.seed(1234)
n = 600; ntrain = 300
p = 20
x = matrix(rnorm(n*p), n, p)
# Treatment assignment
w = rbinom(n, 1, 0.5)
# m^*
m.coefs = c(rep(2,10), rep(0, p-10))
m = x %*% m.coefs
# tau
tau.coefs = runif(p, 0.5, 1)*m.coefs
tau = 1.5*m + x%*%tau.coefs
mu = m + w * tau
y = mu + 10 * rnorm(n)
cat("Signal to noise ratio:", var(mu)/var(y-mu))
#> Signal to noise ratio: 2.301315
# Split into train/test
xtest = x[-(1:ntrain), ]
tautest = tau[-(1:ntrain)]
wtest = w[-(1:ntrain)]
x = x[1:ntrain, ]
y = y[1:ntrain]
w = w[1:ntrain]
# Define training folds
nfolds = 10
foldid = sample(rep(1:10, trunc(nrow(x)/nfolds)+1))[1:nrow(x)]We begin model fitting, starting with our estimate of (the probability of receiving the treatment). To fit , we will also need to record the cross-fitted .
e_fit = cv.glmnet(x, w, foldid = foldid,
family="binomial", type.measure="deviance",
keep = TRUE)
e_hat = e_fit$fit.preval[, e_fit$lambda == e_fit$lambda.1se]
e_hat = 1/(1 + exp(-e_hat))Now, stage 1 of pretraining: fit a model for and record the support. As before, we also record the cross-fitted .
m_fit = cv.glmnet(x, y, foldid = foldid, keep = TRUE)
m_hat = m_fit$fit.preval[, m_fit$lambda == m_fit$lambda.1se]
bhat = coef(m_fit, s = m_fit$lambda.1se)
support = which(bhat[-1] != 0)To fit , we will regress on ; we’ll define them here:
y_tilde = y - m_hat
x_tilde = cbind(as.numeric(w - e_hat) * cbind(1, x))And now, pretraining for . Loop over ; for each , fit a model for using the penalty factor defined by the support of and . We’ll keep track of our CV MSE at each step so that we can choose the that minimizes the MSE.
cv.error = NULL
alphalist = seq(0, 1, length.out = 11)
for(alpha in alphalist){
pf = rep(1/alpha, p)
pf[support] = 1
pf = c(0, pf) # Don't penalize the intercept
tau_fit = cv.glmnet(x_tilde, y_tilde,
foldid = foldid,
penalty.factor = pf,
intercept = FALSE, # already include in x_tilde
standardize = FALSE)
cv.error = c(cv.error, min(tau_fit$cvm))
}
plot(alphalist, cv.error, type = "b",
xlab = expression(alpha),
ylab = "CV MSE",
main = bquote("CV mean squared error as a function of " ~ alpha))
abline(v = alphalist[which.min(cv.error)])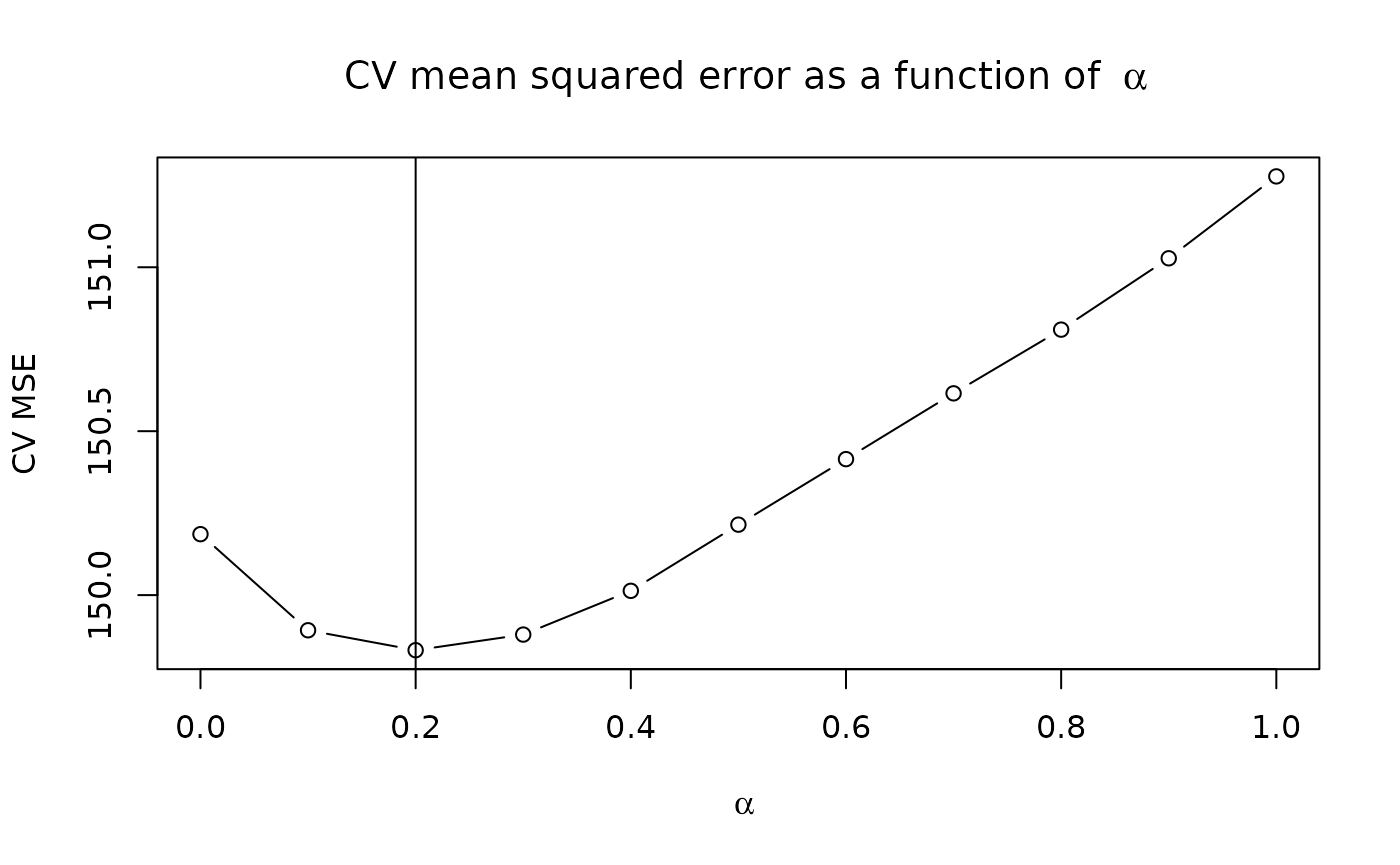
In the plot above, the value at corresponds to the usual R learner, which makes no assumption about a shared support between and . Based on the plot, we choose as our best performing model:
best.alpha = alphalist[which.min(cv.error)]
cat("Chosen alpha:", best.alpha)
#> Chosen alpha: 0.2
pf = rep(1/best.alpha, p)
pf[support] = 1
pf = c(0, pf)
tau_fit = cv.glmnet(x_tilde, y_tilde, foldid = foldid,
penalty.factor = pf,
intercept = FALSE,
standardize = FALSE)To concretely compare the pretrained R-learner with the usual R-learner, we’ll train the usual R-learner here:
tau_rlearner = cv.glmnet(x_tilde, y_tilde, foldid = foldid,
penalty.factor = c(0, rep(1, ncol(x))),
intercept = FALSE,
standardize = FALSE)As anticipated, pretraining improves the prediction squared error relative to the R learner – this is how we designed our simulation:
rlearner_preds = predict(tau_rlearner, cbind(1, xtest), s = "lambda.min")
cat("R-learner PSE: ",
round(mean((rlearner_preds - tautest)^2), 2))
#> R-learner PSE: 45.85
pretrained_preds = predict(tau_fit, cbind(1, xtest), s = "lambda.min")
cat("Pretrained R-learner PSE: ",
round(mean((pretrained_preds - tautest)^2), 2))
#> Pretrained R-learner PSE: 37.63What if the pretraining assumption is wrong?
Here, we repeat everything from above, only now there is no overlap in the support of and .
######################################################
# Simulate data
######################################################
x = matrix(rnorm(n*p), n, p)
# Treatment assignment
w = rbinom(n, 1, 0.5)
# m^*
m.coefs = c(rep(2,10), rep(0, p-10))
m = x %*% m.coefs
# tau
# Note these coefficients have no overlap with m.coefs!
tau.coefs = c(rep(0, 10), rep(2, 10), rep(0, p-20))
tau = x%*%tau.coefs
mu = m + w * tau
y = mu + 10 * rnorm(n)
cat("Signal to noise ratio:", var(mu)/var(y-mu))
#> Signal to noise ratio: 0.6938152
# Split into train/test
xtest = x[-(1:ntrain), ]
tautest = tau[-(1:ntrain)]
wtest = w[-(1:ntrain)]
x = x[1:ntrain, ]
y = y[1:ntrain]
w = w[1:ntrain]
######################################################
# Model fitting: e^*
######################################################
e_fit = cv.glmnet(x, w, foldid = foldid,
family="binomial", type.measure="deviance",
keep = TRUE)
e_hat = e_fit$fit.preval[, e_fit$lambda == e_fit$lambda.1se]
e_hat = 1/(1 + exp(-e_hat))
######################################################
# Model fitting: m^*
######################################################
m_fit = cv.glmnet(x, y, foldid = foldid, keep = TRUE)
m_hat = m_fit$fit.preval[, m_fit$lambda == m_fit$lambda.1se]
bhat = coef(m_fit, s = m_fit$lambda.1se)
support = which(bhat[-1] != 0)
######################################################
# Pretraining: tau
######################################################
y_tilde = y - m_hat
x_tilde = cbind(as.numeric(w - e_hat) * cbind(1, x))
cv.error = NULL
alphalist = seq(0, 1, length.out = 11)
for(alpha in alphalist){
pf = rep(1/alpha, p)
pf[support] = 1
pf = c(0, pf) # Don't penalize the intercept
tau_fit = cv.glmnet(x_tilde, y_tilde,
foldid = foldid,
penalty.factor = pf,
intercept = FALSE, # already include in x_tilde
standardize = FALSE)
cv.error = c(cv.error, min(tau_fit$cvm))
}
# Our final model for tau:
best.alpha = alphalist[which.min(cv.error)]
cat("Chosen alpha:", best.alpha)
#> Chosen alpha: 1
pf = rep(1/best.alpha, p)
pf[support] = 1
pf = c(0, pf)
tau_fit = cv.glmnet(x_tilde, y_tilde, foldid = foldid,
penalty.factor = pf,
intercept = FALSE,
standardize = FALSE)
######################################################
# Fit the usual R-learner:
######################################################
tau_rlearner = cv.glmnet(x_tilde, y_tilde, foldid = foldid,
penalty.factor = c(0, rep(1, ncol(x))),
intercept = FALSE,
standardize = FALSE)
######################################################
# Measure performance:
######################################################
rlearner_preds = predict(tau_rlearner, cbind(1, xtest), s = "lambda.min")
cat("R-learner prediction squared error: ",
round(mean((rlearner_preds - tautest)^2), 2))
#> R-learner prediction squared error: 31.11
pretrained_preds = predict(tau_fit, cbind(1, xtest), s = "lambda.min")
cat("Pretrained R-learner prediction squared error: ",
round(mean((pretrained_preds - tautest)^2), 2))
#> Pretrained R-learner prediction squared error: 31.11Pretraining has not hurt our performance, even though the support of
and
are not shared. Why? Recall that we defined
,
so the relationship between
and
is a function of the supports of both
and
.
In the first stage of pretraining, we fitted
using y ~ x – so the support of
should include the support of
.
As a result, using pretraining with the R-learner should not harm
predictive performance.