Suppose we have a dataset with features and response , and no input grouping. Suppose we also have a small set of meaningful features that we expect to stratify observations (e.g. in biomedicine, may consist of age and sex). In this setting, we can learn input groups using .
The steps to do this are as follows.
- Partition data into two sets: one to learn the grouping and one to do pretraining.
- With the first set, train a small CART tree using and .
- Make predictions for the remaining data; assign observations to groups according to their terminal nodes.
- Apply pretraining using the learned group assignments.
Here, we show an example using simulated data. We use
rpart to train a CART tree. The package ODRF
(Liu and Xia (2022)) is another good
choice – it fits a linear model in each terminal node, which is closer
to what pretraining does, and may therefore have better performance.
#> Loading required package: ptLasso
#> Loading required package: ggplot2
#> Loading required package: glmnet
#> Loading required package: Matrix
#> Loaded glmnet 4.1-10
#> Loading required package: gridExtraSimulate data with a binary outcome: is drawn from a random normal (with uncorrelated features), and is simulated as age (uniform between 20 and 90) and sex (half 0, half 1). The true groups are (1) age under 50, (2) age over 50 and sex = 0 and (3) age over 50 and sex = 1.
set.seed(1234)
n = 1000; p = 50
groupvars = cbind(age = round(runif(n, min = 20, max = 90)),
sex = sample(c(0, 1), n, replace = TRUE))
groups = rep(1, n)
groups[groupvars[, "age"] > 50 & groupvars[, "sex"] == 0] = 2
groups[groupvars[, "age"] > 50 & groupvars[, "sex"] == 1] = 3Now, we’ll define coefficients such that for each group. Across groups, three coefficients are shared, three are group-specific and the rest are 0. Each group has a unique intercept to adjust its baseline risk.
beta.group1 = c(-0.5, 0.5, 0.1, c(0.1, 0.2, 0.3), rep(0, p-6));
beta.group2 = c(-0.5, 0.5, 0.1, rep(0, 3), c(0.1, 0.2, 0.3), rep(0, p-9));
beta.group3 = c(-0.5, 0.5, 0.1, rep(0, 6), c(0.1, 0.2, 0.3), rep(0, p-12));
x = matrix(rnorm(n * p), nrow = n, ncol = p)
x.beta = rep(0, n)
x.beta[groups == 1] = x[groups == 1, ] %*% beta.group1 - 0.75
x.beta[groups == 2] = x[groups == 2, ] %*% beta.group2
x.beta[groups == 3] = x[groups == 3, ] %*% beta.group3 + 0.75
y = rbinom(n, size = 1, prob = 1/(1 + exp(-x.beta)))
# Now that we have our data, we will partition it into 3 datasets:
# one to cluster, one to train models and one to test performance.
xcluster = x[1:250, ]; xtrain = x[251:750, ]; xtest = x[751:1000, ];
ycluster = y[1:250]; ytrain = y[251:750]; ytest = y[751:1000];
zcluster = groupvars[1:250, ];
ztrain = groupvars[251:750, ];
ztest = groupvars[751:1000, ];
# We will use this just to see how our clustering performed.
# Not possible with real data!
groupstrain = groups[251:750]; By design, is different across groups:
ggplot() +
geom_boxplot(aes(x=groups, y=1/(1 + exp(-x.beta)), group = groups)) +
labs(x = "Group", y = "P(y = 1)") +
theme_minimal()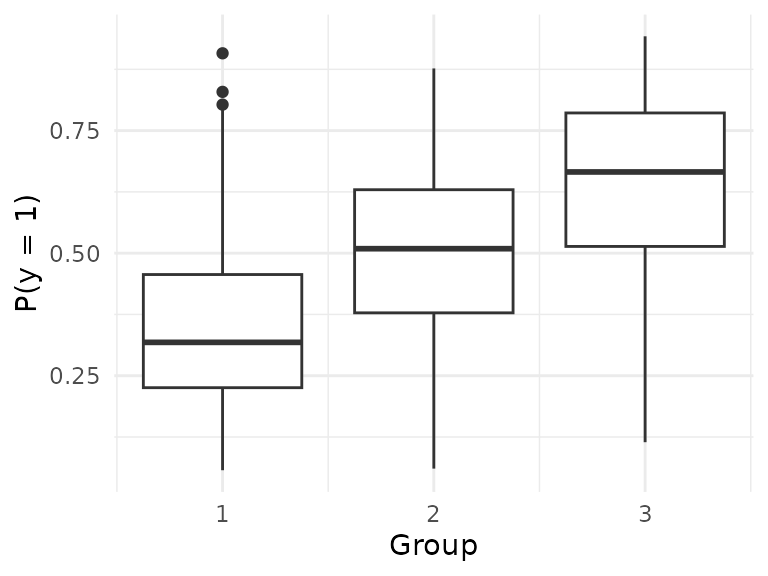
We cluster using rpart. Note that we use
maxdepth = 2: an obvious choice because we simulated the
data and we know that there is a second-level interaction (age + sex)
that determines outcome. In general, however, we recommend keeping this
tree small (maxdepth smaller than 4) so that it is easily
interpretable.
treefit = rpart(ycluster~.,
data = data.frame(zcluster, ycluster),
control=rpart.control(maxdepth=2, minbucket=20))
treefit
#> n= 250
#>
#> node), split, n, deviance, yval
#> * denotes terminal node
#>
#> 1) root 250 61.82400 0.4480000
#> 2) age< 50.5 111 23.18919 0.2972973 *
#> 3) age>=50.5 139 34.10072 0.5683453
#> 6) sex< 0.5 56 13.92857 0.4642857 *
#> 7) sex>=0.5 83 19.15663 0.6385542 *We want our tree to return the ID of the terminal node for each
observation instead of class probabilities. The following is a trick
that causes predict to behave as desired.
leaf=treefit$frame[,1]=="<leaf>"
treefit$frame[leaf,"yval"]=1:sum(leaf)
predgroupstrain = predict(treefit, data.frame(ztrain))
predgroupstest = predict(treefit, data.frame(ztest))Finally, we are ready to apply pretraining using the predicted groups as our grouping variable.
cvfit = cv.ptLasso(xtrain, ytrain, predgroupstrain, family = "binomial",
type.measure = "auc", nfolds = 3,
overall.lambda = "lambda.min")
predict(cvfit, xtest, predgroupstest, ytest = ytest)
#>
#> Call:
#> predict.cv.ptLasso(object = cvfit, xtest = xtest, groupstest = predgroupstest,
#> ytest = ytest)
#>
#>
#> alpha = 0.6
#>
#> Performance (AUC):
#>
#> allGroups mean wtdMean group_1 group_2 group_3
#> Overall 0.7049 0.6508 0.6454 0.6134 0.6722 0.6669
#> Pretrain 0.7113 0.6538 0.6473 0.5979 0.6492 0.7143
#> Individual 0.7097 0.6514 0.6464 0.6085 0.6474 0.6983
#>
#> Support size:
#>
#> Overall 5
#> Pretrain 6 (5 common + 1 individual)
#> Individual 20Note that the overall model trained by cv.ptLasso takes
advantage of the clustering: it fits a unique intercept for each group.
Performance would have been much worse if we hadn’t done any clustering
at all:
baseline.model = cv.glmnet(xtrain, ytrain, family = "binomial", type.measure = "auc", nfolds = 5)
assess.glmnet(baseline.model, newx=xtest, newy=ytest)$auc
#> [1] 0.6130757
#> attr(,"measure")
#> [1] "AUC"